Xie Research Group: Epigenetics and Computational Laboratory

Our laboratory is using neuron stem cells and primary neuronal cultures for functional assays to understand the epigenetic mechanisms underlying brain development and neurological disorders. Transgenic mouse models are exploited to understand how brain cells gain specific functions and properties in response to environmental stimuli.
In addition, our lab has implemented a number of computational pipelines to perform high-throughput sequencing data analysis for RNAseq, ChIPseq, methylome sequencing and single cell “Omics” data.
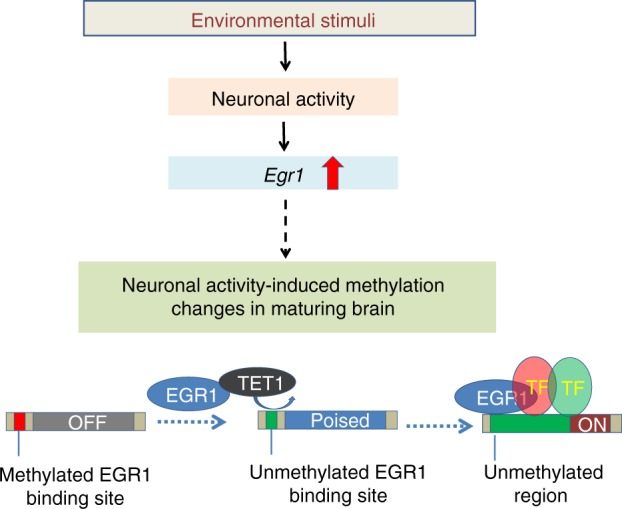
Life experiences can leave lasting marks in the brain, such as epigenetic changes in neurons induced by neuronal activity. How one's life experience or neuronal activity is translated into storable epigenetic information remains largely unknown. Recently, I have been studying the transcriptional regulatory networks associated with brain development and neuronal activity. This body of work discusses how transcription factors may direct brain epigenome programming including histone modifications and DNA methylation.
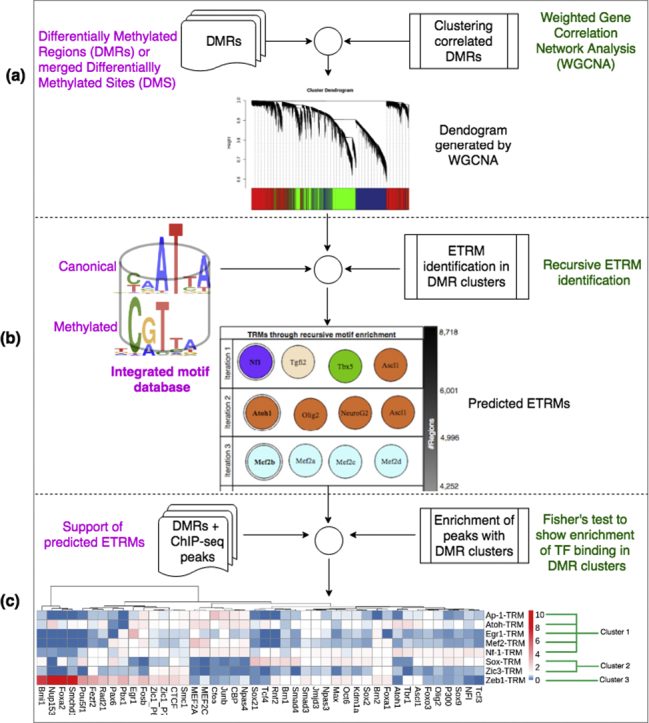
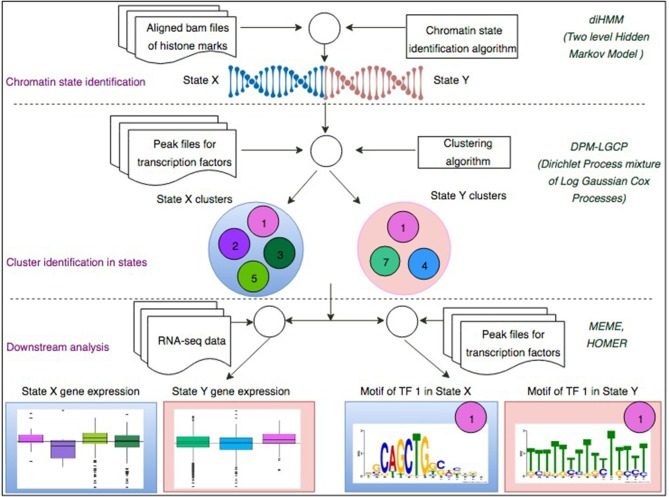
Our studies integrating systems biology with neuroscience emphasize the cross-talks among histone modifications, transcription factor bindings, and DNA methylation. We have implemented a number of computational pipelines: (1) to partition the genome into chromatin states according to histone marks and the distributions of transcription factor binding; (2) to predict transcription regulatory modules via recursive motif search on epigenetic regulated loci; (3) to virtually dissect methylome using single cell data; (4) to predict epigenetic regulatory modules controlling brain cell specification via the integration of single cell “omics” data.
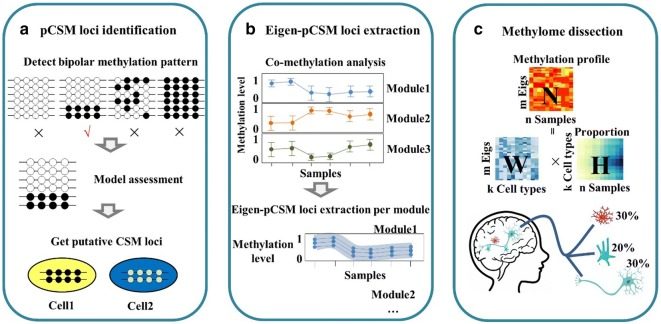
Traditionally, DNA methylation data analysis is based on the determination of the average methylation level (the percentage of methylated CpG) of one or more contiguous CpG sites. However, this conventional way of data analysis is unable to dissect epigenetic heterogeneity associated with distinct mechanisms such as asymmetric DNA methylation, allelic-specific DNA methylation, and cell-type specific DNA methylation. We have developed a nonparametric bayesian clustering approach to detect bipolar methylated genomic loci and implemented a computational pipeline to decipher bipolar methylation patterns and to infer cell-type specific methylated loci. These studies on methylation variation have led to open source tools that will help to interpret epigenetic heterogeneity and will facilitate the modeling of the cell population dynamics at the epigenetic level during normal development and diseases.
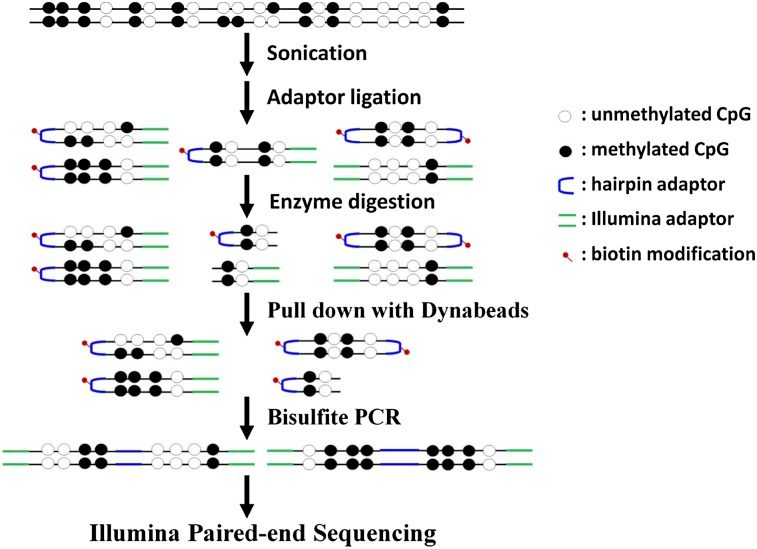
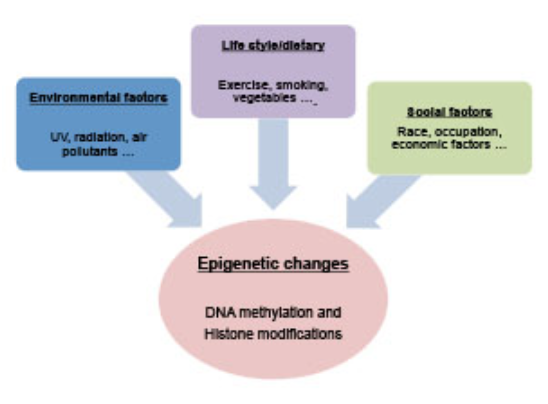
In an effort to understand Genome-Environment interactions, I have, along with my collaborators, determined epigenetic aberrations associated with various kinds of environmental diseases. Our study on pediatric patients has shed light on how environmental factors may alter normal developmental process via critical transcription factors. We have also determined the individual variation and longitudinal pattern of blood methylome during early postnatal human development. Furthermore, we have also studied how environmental factors, including pesticide exposure, may shape the human epigenome.
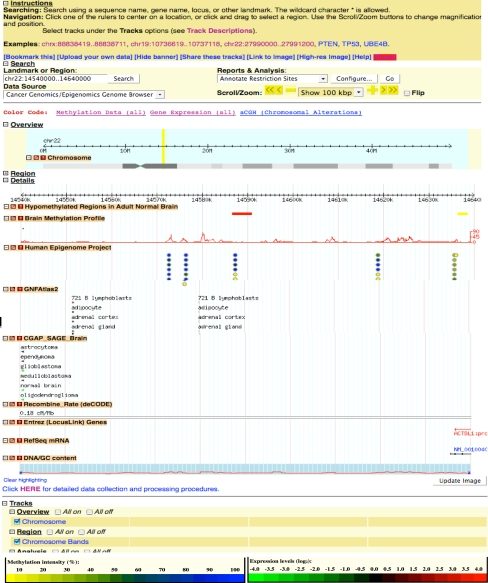
My early publications were focused on the analyses of the genome, transcriptome, and epigenome of developing brains and brain tumors. I have implemented genome-wide Alu-anchored bisulfite sequencing technique to assess the methylation patterns of Alu repeats. This approach facilitates the analysis of methylation changes on sequences of low complexity, such as the bisulfite converted genome, and the studies of global methylation loss, which is frequently observed in tumors. I have also exploited microarray and high-throughput sequencing for gene expression analyses to uncover gene/protein networks whose functions are consistently compromised in brain tumors.
Lab Members
|
Name
|
Title
|
|---|---|
|
Alajoleen, Razan |
Visiting Student |
|
Armstrong, Nicole |
BREU Student |
|
Fan, Jiayi |
Postdoc |
|
Johnson, Zachary |
Graduate Research Assistant |
|
Melville, Natalie |
Graduate Research Assistant |
|
Murray, Alexander |
Graduate Research Assistant |
|
Wei, Xiaoran |
Visiting Student |
|
Xu, Xiguang |
Visiting Student |
Featured Publications
Sun Z., Xu X., He J., Murray A., Sun M., Wei X., Wang X., McCoig E., Xie E., Xi J., Li L., Zhu J., Chen J., Morozov A., Pickrell A.M., Theus M.H. and Xie, H. (2019) EGR1 Recruits TET1 to Shape the Brain Methylome during Development and upon Neuronal Activity, Nature Communications, 10(1):3892. PMID:31467272 https://www.ncbi.nlm.nih.gov/pubmed/31467272.
He J., Xu X., Monavarfeshani A., Banerjee S., Fox A. M., Xie H. (2019) Retinal Input Induced Epigenetic Dynamics in the Developing Mouse Dorsal Lateral Geniculate Nucleus, Epigenetics & Chromatin;12(1):13. PMID:30764861 https://www.ncbi.nlm.nih.gov/pubmed/30764861.
Zhao L, Sun M., Li Z., Bai X., Yu M., Wang M., Liang L., Shao X., Arnovitz S., Wang Q., He C., Lu X., Chen J., Xie H. (2014) The dynamics of DNA methylation fidelity during mouse embryonic stem cell self-renewal and differentiation. Genome Research. 24(8):1296-307. PMID: 24835587. PMCID: PMC4120083 https://www.ncbi.nlm.nih.gov/pubmed/24835587.
Xie H., Wang M., Bonaldo M. F., Rajaram V., Stellpflug W., Smith C., Arndt K., Goldman S., Tomita T., and Soares M. B. (2010) Epigenomic analysis of Alu repeats in human ependymomas. Proc Natl Acad Sci U S A, 107(15):6952-7. PMID: 20351280. PMCID: PMC2872440 https://www.ncbi.nlm.nih.gov/pubmed/20351280.
2019
Rahtes A., Pradhan K., Sarma M., Xie D., Lu C., Li L. (2019) Phenylbutyrate facilitates homeostasis of non-resolving inflammatory macrophages. Innate Immunity. PMID: 31604378. https://www.ncbi.nlm.nih.gov/pubmed/31604378.
Zhang M, Wang J, Zhang K, Lu G, Xu L, Ren K, Liu Y, Xing J, Gao X, Jin W, Berry K, Xie H, Wu S, Lu Q, Zhao X. TET1-mediated Oligodendrocyte Homeostasis Regulates Myelination and Synaptic Functions. 2019 https://www.biorxiv.org/content/10.1101/821496v1.
Yin L, Luo Y, Xu X, Wen S, Wu X, Lu X, Xie H. Virtual methylome dissection facilitated by single-cell analyses. Epigenetics & Chromatin 12, 66 (2019) https://doi.org/10.1186/s13072-019-0310-9
Xu X, Wei X, Xie H. Advances in methods and software for RNA cytosine methylation analysis. Genomics. 2019 Oct 30. pii: S0888-7543(19)30516-6. https://doi.org/10.1016/j.ygeno.2019.10.017
Sun Z, Xu X, He J, Murray A, Sun MA, Wei X, Wang X, McCoig E, Xie E, Jiang X, Li L, Zhu J, Chen J, Morozov A, Pickrell AM, Theus MH, Xie H. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat Commun. 2019 Aug 29;10(1):3892. https://doi.org/10.1038/s41467-019-11905-3
Hazy A, Bochicchio L, Oliver A, Xie E, Geng S, Brickler T, Xie H, Li L, Allen IC, Theus MH. Divergent age-dependent peripheral immune transcriptomic profile following traumatic brain injury. Sci Rep. 2019 Jun 12;9(1):8564. https://doi.org/10.1038/s41598-019-45089-z
Banerjee S, Wei X, Xie H. Recursive Motif Analyses Identify Brain Epigenetic Transcription Regulatory Modules. Comput Struct Biotechnol J. 2019 Apr 9;17:507-515. https://doi.org/10.1016/j.csbj.2019.04.003
He J, Xu X, Monavarfeshani A, Banerjee S, Fox MA, Xie H. Retinal-input-induced epigenetic dynamics in the developing mouse dorsal lateral geniculate nucleus. Epigenetics Chromatin. 2019 Feb 14;12(1):13. https://doi.org/10.1186/s13072-019-0257-x
Banerjee S, Zhu H, Tang M, Feng WC, Wu X, Xie H. Identifying Transcriptional Regulatory Modules Among Different Chromatin States in Mouse Neural Stem Cells. Front Genet. 2019 Jan 15;9:731. https://doi.org/10.3389/fgene.2018.00731
2018
Ma S, de la Fuente Revenga M, Sun Z, Sun C, Murphy TW, Xie H, González-Maeso J, Lu C. Cell-type-specific brain methylomes profiled via ultralow-input microfluidics. Nat Biomed Eng. 2018 Mar;2(3):183-194. https://doi.org/10.1038/s41551-018-0204-3
Luo Y, He J, Xu X, Sun MA, Wu X, Lu X, Xie H. Integrative single-cell omics analyses reveal epigenetic heterogeneity in mouse embryonic stem cells. PLoS Comput Biol. 2018 Mar 21;14(3):e1006034. https://doi.org/10.1371/journal.pcbi.1006034
Oestreich K, Xie H. Interviewed by Tech V. Scientist awarded $2 million grant from the NIH to study the body's immune memory response. EurekAlert. 2018. https://www.eurekalert.org/pub_releases/2018-06/vt-sa062718.php
2017
Banjeree S, Chem X, Wu X, Xie H, Xuan J, Feng WC. ChIP-GMM: A Gaussian mixture model for inferring binding regions in ChIP-seq proffles. Proceedings of the 9th International Conference on Bioinformatics and Computational Biology, BICOB 2017. 2017. https://loop.frontiersin.org/publications/48763699.
2016
Tran H, Wu X, Tithi S, Sun M-A, Xie H, Zhang L. A Bayesian Assignment Method for Ambiguous Bisulfite Short Reads. PLOS ONE. 2016;11(3). https://doi.org/10.1371/journal.pone.0151826.
Sharif J, Endo TA, Nakayama M, Karimi MM, Shimada M, Katsuyama K, Goyal P, Brind’Amour J, Sun M-A, Sun Z, Ishikura T, Mizutani-Koseki Y, Ohara O, Shinkai Y, Nakanishi M, Xie H, Lorincz MC, Koseki H. Activation of Endogenous Retroviruses in Dnmt1 −/− ESCs Involves Disruption of SETDB1-Mediated Repression by NP95 Binding to Hemimethylated DNA. Cell Stem Cell. 2016;19(1):81–94. https://doi.org/10.1016/j.stem.2016.03.013.
Okyere B, Giridhar K, Hazy A, Chen M, Keimig D, Bielitz RC, Xie H, He J-Q, Huckle WR, Theus MH. Endothelial-Specific EphA4 Negatively Regulates Native Pial Collateral Formation and Re-Perfusion following Hindlimb Ischemia. PLOS ONE. 2016;11(7). https://doi.org/10.1371/journal.pone.0159930.
von Meyenn F, Iurlaro M, Habibi E, Liu NQ, Salehzadeh-Yazdi A, Santos F, Petrini E, Milagre I, Yu M, Xie Z, Kroeze LI, Nesterova TB, Jansen JH, Xie H, He C, Reik W, Stunnenberg HG. Impairment of DNA Methylation Maintenance Is the Main Cause of Global Demethylation in Naive Embryonic Stem Cells. Molecular Cell. 2016;62(6):848–861. https://doi.org/10.1016/j.molcel.2016.04.025.
Sun M-A, Sun Z, Wu X, Rajaram V, Keimig D, Lim J, Zhu H, Xie H. Mammalian Brain Development is Accompanied by a Dramatic Increase in Bipolar DNA Methylation. SCIENTIFIC REPORTS. 2016;6. https://doi.org/10.1038/srep32298.
2015
Porter J, Sun M-A, Xie H, Zhang L. Investigating bisulfite short-read mapping failure with hairpin bisulfite sequencing data. Presented at the 4th IEEE International Conference on Computational Advances in Bio and Medical Sciences (ICCABS), Miami Beach, FL; 2015. https://doi.org/10.1186/1471-2164-16-S11-S2
He J, Sun M-A, Wang Z, Wang Q, Li Q, Xie H. Characterization and machine learning prediction of allele-specific DNA methylation. GENOMICS. 2015;106(6):331–339. https://doi.org/10.1016/j.ygeno.2015.09.007
Wu X, Sun M-A, Zhu H, Xie H. Nonparametric Bayesian clustering to detect bipolar methylated genomic loci. BMC BIOINFORMATICS. 2015;16. https://doi.org/10.1186/s12859-014-0439-2
Sun M-A, Velmurugan KR, Keimig D, Xie H. HBS-Tools for Hairpin Bisulfite Sequencing Data Processing and Analysis. Advances in Bioinformatics. 2015;2015:1–4. https://doi.org/10.1155/2015/760423
2014
Zhao L, Sun M-A, Li Z, Bai X, Yu M, Wang M, Liang L, Shao X, Arnovitz S, Wang Q, He C, Lu X, Chen J, Xie H. The dynamics of DNA methylation fidelity during mouse embryonic stem cell self-renewal and differentiation. GENOME RESEARCH. 2014;24(8):1296–1307. https://doi.org/10.1101/gr.163147.113.
Shao X, Zhang C, Sun M-A, Lu X, Xie H. Deciphering the heterogeneity in DNA methylation patterns during stem cell differentiation and reprogramming. BMC Genomics. 2014;15(1):978–978. https://doi.org/10.1186/1471-2164-15-978.
Luo Y, Lu X, Xie H. Dynamic Alu methylation during normal development, aging, and tumorigenesis. BioMed research international. 2014;2014:784706. https://doi.org/10.1155/2014/784706.
McErlean P, Favoreto S, Costa FF, Shen J, Quraishi J, Biyasheva A, Cooper JJ, Scholtens DM, Vanin EF, Bonaldo MF de, Xie H, Soares MB, Avila PC. Human rhinovirus infection causes different DNA methylation changes in nasal epithelial cells from healthy and asthmatic subjects. BMC Medical Genomics. 2014;7(1). https://doi.org/10.1186/1755-8794-7-37.
Tran H, Porter J, Sun M-A, Sun M-A, Xie H, Zhang L. Objective and comprehensive evaluation of bisulfite short read mapping tools. Advances in bioinformatics. 2014;2014:472045. https://doi.org/10.1155/2014/472045
2013
He J, Sun X, Shao X, Liang L, Xie H. DMEAS: DNA methylation entropy analysis software. Bioinformatics. 2013;29(16):2044–2045. https://doi.org/10.1093/bioinformatics/btt332
2012
Hoxha E, Lambers E, Xie H, De Andrade A, Krishnamurthy P, Wasserstrom JA, Ramirez V, Thal M, Verma SK, Soares MB, Kishore R. Histone Deacetylase 1 Deficiency Impairs Differentiation and Electrophysiological Properties of Cardiomyocytes Derived from Induced Pluripotent Cells. STEM CELLS. 2012;30(11):2412–2422. https://doi.org/10.1002/stem.1209.
Wang M, Xie H, Shrestha S, Sredni S, Morgan GA, Pachman LM. Methylation alterations of WT1 and homeobox genes in inflamed muscle biopsy samples from patients with untreated juvenile dermatomyositis suggest self-renewal capacity. Arthritis & Rheumatism. 2012;64(10):3478–3485. https://doi.org/10.1002/art.34573.
Zhang X, Wallace AD, Du P, Kibbe WA, Jafari N, Xie H, Lin S, Baccarelli A, Soares MB, Hou L. DNA methylation alterations in response to pesticide exposure in vitro. Environmental and Molecular Mutagenesis. 2012;53(7):542–549. https://doi.org/10.1002/em.21718.
Principal Investigator

Hehuang (David) Xie, PhD
Associate Professor
Fralin Life Sciences Institute (MC 0477)
Steger Hall | Office 263D
1015 Life Science Circle
Blacksburg, VA 24061-0477
Email: davidxie@vt.edu
Office Phone: (540) 231-9244
Lab Phone: (540) 231-6128


